Szalai, Tibor Viktor and Péczka, Nikolett and Sipos-Szabó, Levente and Petri, László and Bajusz, Dávid and Keserű, György Miklós (2025) Ultrahigh-Throughput Virtual Screening Strategies against PPI Targets: A Case Study of STAT Inhibitors. JOURNAL OF CHEMICAL INFORMATION AND MODELING, 65 (14). pp. 7734-7748. ISSN 1549-9596
|
Text
szalai-et-al-2025-ultrahigh-throughput-virtual-screening-strategies-against-ppi-targets-a-case-study-of-stat-inhibitors.pdf - Published Version Available under License Creative Commons Attribution. Download (10MB) | Preview |
|
![[img]](https://real.mtak.hu/224477/7.hassmallThumbnailVersion/graphicalabs.jpeg)
|
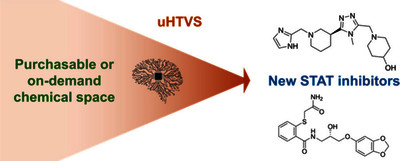
Text (graphical abstract)
graphicalabs.jpeg - Published Version Available under License Creative Commons Attribution. Download (81kB) | Preview |
Abstract
In recent years, virtual screening of ultralarge (108+) libraries of synthetically accessible compounds (uHTVS) became a popular approach in hit identification. With AI-assisted virtual screening workflows, such as Deep Docking, these protocols might be feasible even without supercomputers. Yet, these methodologies have their own conceptual limitations, including the fact that physics-based docking is replaced by a cheaper deep learning (DL) step for the vast majority of compounds. In turn, the performance of this DL step will highly depend on the performance of the underlying docking model that is used to evaluate parts of the whole data set to train the DL architecture itself. Here, we evaluated the performance of the popular Deep Docking workflow on compound libraries of different sizes, against benchmark cases of classic brute-force docking approaches conducted on smaller libraries. We were especially interested in more difficult, protein-protein interaction-type oncotargets where the reliability of the underlying docking model is harder to assess. Specifically, our virtual screens have resulted in several new inhibitors of two oncogenic transcription factors, STAT3 and STAT5b. For STAT5b, in particular, we disclose the first application of virtual screening against its N-terminal domain, whose importance was recognized more recently. While the AI-based uHTVS is computationally more demanding, it can achieve exceptionally good hit rates (50.0% for STAT3). Deep Docking can also work well with a compound library containing only several million (instead of several billion) compounds, achieving a 42.9% hit rate against the SH2 domain of STAT5b, while presenting a highly economic workflow with just under 120,000 compounds actually docked. © 2025 The Authors. Published by American Chemical Society.
| Item Type: | Article |
|---|---|
| Additional Information: | Acknowledgments: The authors thank Elvin de Araujo and Qirat Ashraf (University of Toronto) for providing the STAT proteins for the experimental work. This work was supported by the National Research Development and Innovation Office of Hungary [contracts FK146063 to D.B., K135150 and PharmaLab (RRF-2.3.1-21-2022-00015) to G.M.K.]. The work of D.B. was supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences. We acknowledge the HPC time and support of the Governmental Information Technology Development Agency, Hungary. |
| Subjects: | R Medicine / orvostudomány > R1 Medicine (General) / orvostudomány általában |
| SWORD Depositor: | MTMT SWORD |
| Depositing User: | MTMT SWORD |
| Date Deposited: | 17 Sep 2025 19:40 |
| Last Modified: | 17 Sep 2025 19:40 |
| URI: | https://real.mtak.hu/id/eprint/224477 |
Actions (login required)
 |
Edit Item |