Kwan, Alan C. and Chang, Ernest W. and Jain, Ishan and Theurer, John and Tang, Xiu and Francisco, Nadia and Haddad, Francois and Liang, David and Fábián, Alexandra and Ferencz, Andrea and Yuan, Neal and Merkely, Béla and Siegel, Robert and Cheng, Susan and Kovács, Attila and Tokodi, Márton and Ouyang, David (2024) Deep Learning-Derived Myocardial Strain. JACC: Cardiovascular Imaging, 17 (7). pp. 715-725. ISSN 1936878X
|
Text
kwan-et-al-2024-deep-learning-derived-myocardial-strain.pdf - Published Version Download (5MB) | Preview |
|
![[img]](https://real.mtak.hu/224868/2.hassmallThumbnailVersion/1-s2.0-S1936878X24000639-ga1_lrg333.jpg)
|
Text (graphical abstract)
1-s2.0-S1936878X24000639-ga1_lrg333.jpg - Published Version Download (1MB) | Preview |
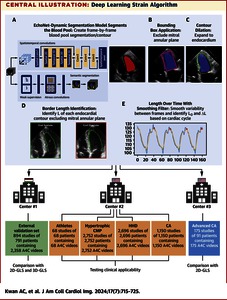
Abstract
Background Echocardiographic strain measurements require extensive operator experience and have significant intervendor variability. Creating an automated, open-source, vendor-agnostic method to retrospectively measure global longitudinal strain (GLS) from standard echocardiography B-mode images would greatly improve post hoc research applications and may streamline patient analyses. Objectives This study was seeking to develop an automated deep learning strain (DLS) analysis pipeline and validate its performance across multiple applications and populations. Methods Interobserver/-vendor variation of traditional GLS, and simulated effects of variation in contour on speckle-tracking measurements were assessed. The DLS pipeline was designed to take semantic segmentation results from EchoNet-Dynamic and derive longitudinal strain by calculating change in the length of the left ventricular endocardial contour. DLS was evaluated for agreement with GLS on a large external dataset and applied across a range of conditions that result in cardiac hypertrophy. Results In patients scanned by 2 sonographers using 2 vendors, GLS had an intraclass correlation of 0.29 (95% CI: −0.01 to 0.53, P = 0.03) between vendor measurements and 0.63 (95% CI: 0.48-0.74, P < 0.001) between sonographers. With minor changes in initial input contour, step-wise pixel shifts resulted in a mean absolute error of 3.48% and proportional strain difference of 13.52% by a 6-pixel shift. In external validation, DLS maintained moderate agreement with 2-dimensional GLS (intraclass correlation coefficient [ICC]: 0.56, P = 0.002) with a bias of −3.31% (limits of agreement: −11.65% to 5.02%). The DLS method showed differences (P < 0.0001) between populations with cardiac hypertrophy and had moderate agreement in a patient population of advanced cardiac amyloidosis: ICC was 0.64 (95% CI: 0.53-0.72), P < 0.001, with a bias of 0.57%, limits of agreement of −4.87% to 6.01% vs 2-dimensional GLS. Conclusions The open-source DLS provides lower variation than human measurements and similar quantitative results. The method is rapid, consistent, vendor-agnostic, publicly released, and applicable across a wide range of imaging qualities.
| Item Type: | Article |
|---|---|
| Uncontrolled Keywords: | deep learning, echocardiography, longitudinal strain |
| Subjects: | R Medicine / orvostudomány > RC Internal medicine / belgyógyászat |
| Depositing User: | Dr. Attila Kovács |
| Date Deposited: | 22 Sep 2025 13:18 |
| Last Modified: | 22 Sep 2025 13:18 |
| URI: | https://real.mtak.hu/id/eprint/224868 |
Actions (login required)
 |
Edit Item |